 |
 |
Section 6: Visualization
Assume we have used the dataloader to, e.g., learn a generative model on random patches. Assume we want to “see” the patches that the dataloader has returned. As in section 2, the code for training is rougly as follows:
dataloader.start()
time.sleep(10) #wait for the dataloader to load initial BigChunks.
while True:
x, list_patients, list_smallchunks = dataloader.get()
'''
`x` is now a tensor of shape [`batch_size` x 3 x 224 x 224].
`list_patients` is a list of lenght `batch_size`.
TODO: You can use these values to, e.g., update model parameters.
.
.
.
'''
if(flag_finish_running == True):
dataloader.pause_loading()
break
To visualize the collected SmallChunks, you only need to implement a function that takes in:
patient: an instnace ofPatient.list_smallchunks: a list ofSmallChunks. This list contains all the returnedSmallChunks that correspond topatient.
Here is an example implementation of such a function.
This function shows a thumbnail of the patient’s H&E slide. Afterwards, it shows BigChunks as squares and SmallChunks’ centers as points on the thumbnail.
def func_visualize_one_patient(patient, list_smallchunks):
'''
Given all smallchunks collected for a specific patient, this function
should visualize the patient.
Inputs:
- patient:
the patient under considerations, an instance of `utils.data.Patient`.
- list_smallchunks: the list of all collected small chunks for the patient,
a list whose elements are an instance of `lightdl.SmallChunk`.
'''
#settings =======
vis_scale = 0.1 #=====
fname_wsi = patient.dict_records["wsi"].rootdir +\
patient.dict_records["wsi"].relativedir
opsimage = openslide.OpenSlide(fname_wsi)
W, H = opsimage.dimensions
opsimageW, opsimageH = opsimage.dimensions
W, H = int(W*vis_scale), int(H*vis_scale)
pil_thumbnail = opsimage.get_thumbnail((W,H))
plt.ioff()
fig, ax = plt.subplots(1,2, figsize=(2*10,10))
ax[0].imshow(pil_thumbnail) #show the thumbnail
ax[0].axis('off')
ax[0].set_title("patient {}, H&E [{} x {}]."\
.format(patient.int_uniqueid, opsimageW, opsimageH))
ax = ax[1]
ax.imshow(pil_thumbnail)
ax.axis('off')
print("patient {}, number of smallchunks={}"\
.format(patient, len(list_smallchunks)))
list_colors = ['lawngreen', 'cyan', 'gold', 'greenyellow']
list_shownbigchunks = []
for smallchunk in list_smallchunks:
#show the bigchunk ================
x = smallchunk.dict_info_of_bigchunk["x"]
y = smallchunk.dict_info_of_bigchunk["y"]
x, y = int(x*vis_scale), int(y*vis_scale)
if(not([x,y] in list_shownbigchunks)):
w, h = int(1000*vis_scale), int(1000*vis_scale)
rect = patches.Rectangle(
(x,y), w, h, linewidth=1,\
linestyle="--",\
edgecolor=random.choice(list_colors),\
facecolor='none', fill=False
)
ax.add_patch(rect)
list_shownbigchunks.append([x,y])
#get x,y,w,h ======
x = smallchunk.dict_info_of_smallchunk["x"]*vis_scale +\
smallchunk.dict_info_of_bigchunk["x"]*vis_scale
y = smallchunk.dict_info_of_smallchunk["y"]*vis_scale +\
smallchunk.dict_info_of_bigchunk["y"]*vis_scale
x, y = int(x), int(y)
w, h = int(224*vis_scale), int(224*vis_scale)
x_centre, y_centre = int(x+0.5*w), int(y+0.5*h)
#make-show the rect =====
circle = patches.Circle((x_centre, y_centre), radius=w*0.05,\
facecolor=random.choice(list_colors),\
fill=True)
ax.add_patch(circle)
plt.title("patient {} (extracted big/small chunks)"\
.format(patient.int_uniqueid), fontsize=20)
plt.savefig("Visualization/patient_{}.eps"\
.format(patient.int_uniqueid), bbox_inches='tight',\
format='eps')
plt.close(fig)
Once data loading is finished (i.e. after calling dataloader.pause_loading()) we can visualize the collected SmallChunks as follows:
dataloader.visualize(func_visualize_one_patient)
Afterwards, according to the above func_visualize_one_patient for each patient one image will be created.
Here are some sample images. Please note that these are all of the SmallChunks which are returned by the function dataloader.get().
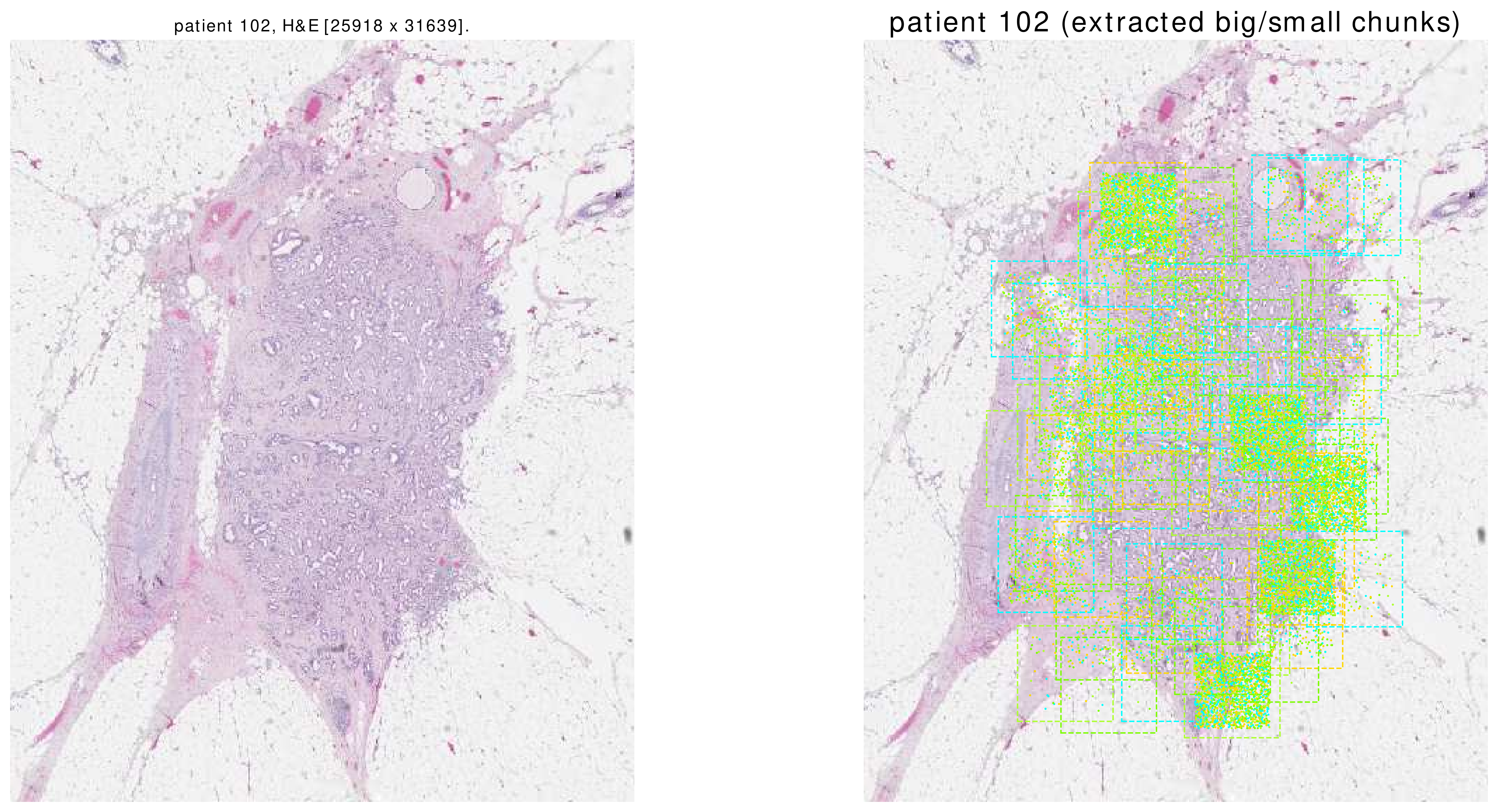
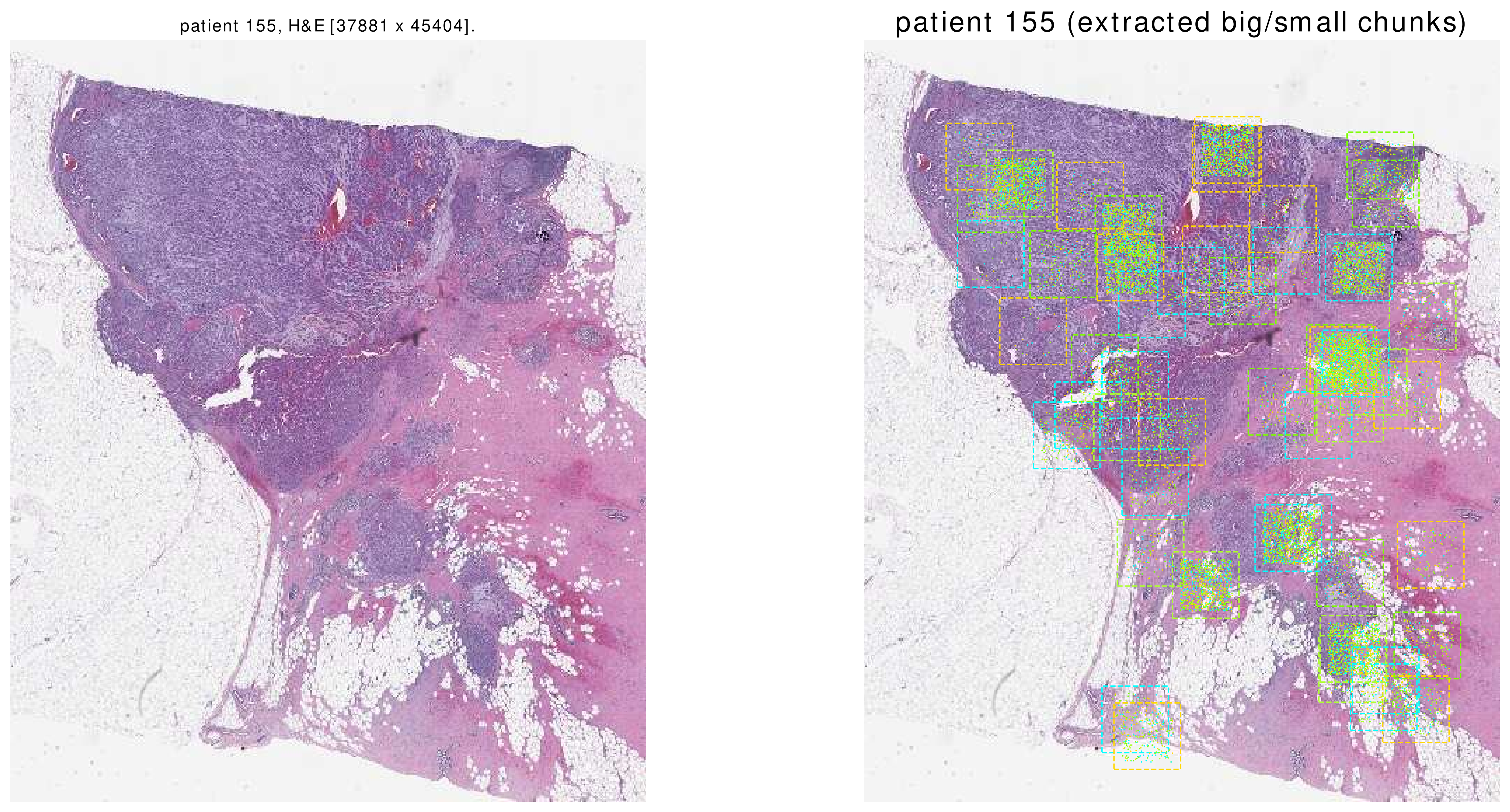
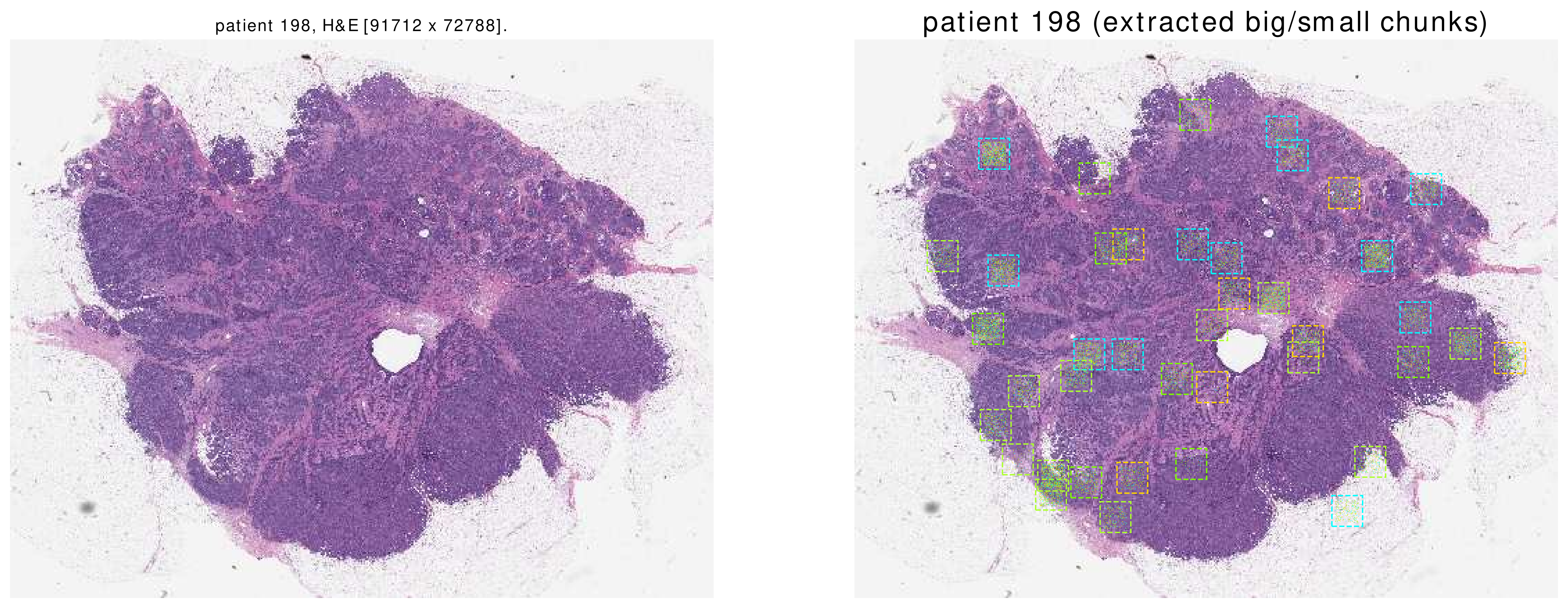
 |
 |