 |
 |
Section 5: Getting Heatmaps for WSIs
Sample notebook to get heatmap is available at the following link: Link to sample notebook.
In this section we explain how to efficiently get output maps (e.g. output activations of a CNN) for huge whole-slide-images in PyDmed. In doing so, we introduce two tools in different subsections:
- Sliding window dataloader, implemented by the class
pydmed.extensions.wsi.SlidingWindowDL. - Collecting and processing a stream of smallchunks.
Sliding-window dataloader
Sliding-window dataloader considers a square of a specific size (a square of size kernel_size), and slides
it over a whole-slide-image with a specific stride as set by the variable stride.
The below figure illustrates how the sliding-window dataloader returns patches.
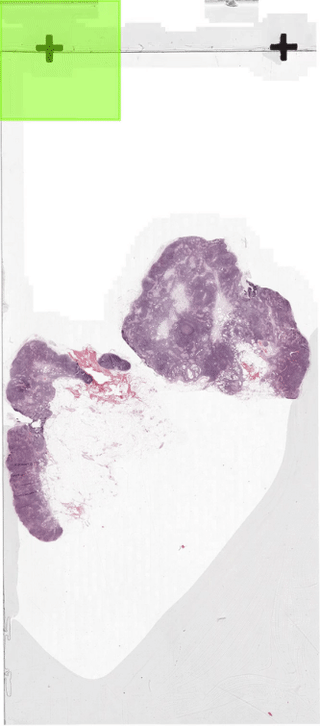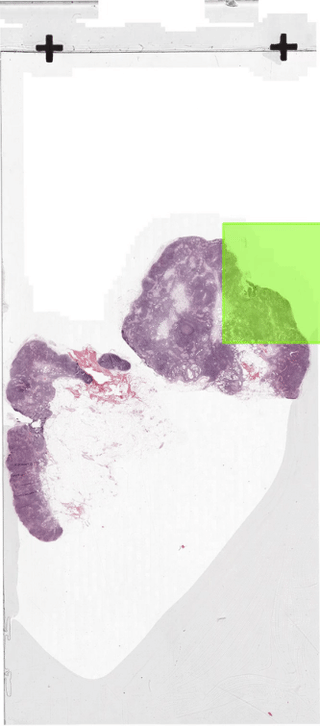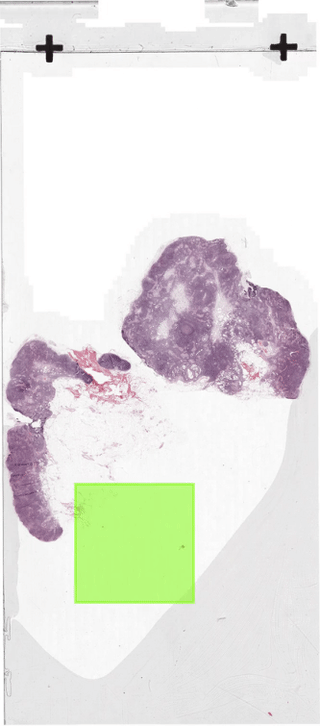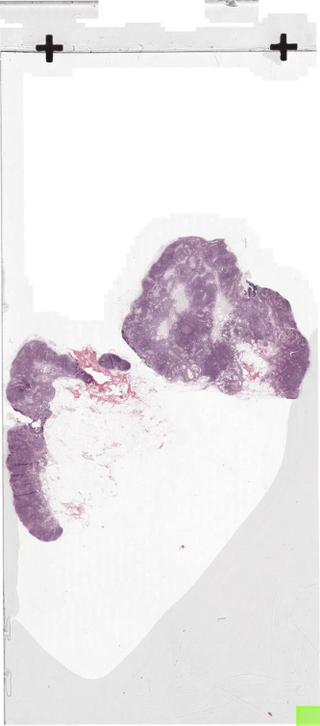
In section 2 we discussed how to create a dataloader.
Making a sliding-window dataloader is almost the same.
The only difference is that sliding-window dataloader (i.e., SlidingWindowDL)
needs few additional arguements. The below code illustrates how to create a sliding-window dataloader:
dl_forheatmap = pydmed.extensions.wsi.SlidingWindowDL(
intorfunc_opslevel = 1,
kernel_size = kernel_size,
stride = kernel_size,
mininterval_loadnewbigchunk = 15,
dataset = dataset,\
type_bigchunkloader=pydmed.extensions.wsi.SlidingWindowBigChunkLoader,\
type_smallchunkcollector=pydmed.extensions.wsi.SlidingWindowSmallChunkCollector,\
const_global_info=const_global_info,\
batch_size=1,\
tfms_onsmallchunkcollection=tfms_onsmallchunkcollection,\
tfms = tfms_oncolate,
flag_grabqueue_onunsched = True
)
The SlidingWindowDL has 4 additional arguments compared to the normal dataloader we discussed in
section 2:
arg1. intorfunc_opslevel: This arguments specifies the pyramidal image’s level from which the patches
are going to be extracted. For example when this argument is set to 1, patches will be extracted from level 1.
In histopathology datasets it is quite common to have slides which are scanned at different magnification levels.
For instnace in a dataset some of the slides might be scanned at 20x magnification level, while some others at 40x magnification level.
In this case, one may need to consider the level based on magnification level (e.g. level 0 for 40x slides and level 1 for 20x slides).
For doing so, you can implement a function that decides a specific level for each WSI. Afterwards, you need to
pass the function as the argument intorfunc_opslevel. The below code demonstrates the procedure:
def func_getopslevel(patient_input):
flag_some_condition_on_patient = ... #Here you can make a boolean.
if(flag_some_condition_on_patient == True):
target_level = 0
else:
target_level = 1
return target_level
dl_forheatmap = pydmed.extensions.wsi.SlidingWindowDL(
intorfunc_opslevel = func_getopslevel,
.... other arguements ....
)
arg2. kernel_size: an integer, the width of the sliding-window. This argument is analogous to
the kernel_size argument of, e.g., pytorch’s Conv2d module.
arg3. stride: an integer, the amount of shift of the sliding-window in each step.
This argument is analogous to
the stride argument of, e.g., pytorch’s Conv2d module.
arg4. mininterval_loadnewbigchunk: the minimum time (in seconds) between loading two BigChunks.
The value is typically between 10 and 30 based on the mahcine being used.
In case you noticed the dataloader is demanding too much from the machine, you can increase this argument.
Processing a stream of small-chunnks
So far we saw how SlidingWindowDL makes a stream of SmallChunks.
But how to process these smallchunks and produce, e.g., a heatmap?
For huge whole-slide-images, heatmaps also become very huge, and it is not often possible to store them in RAM.
Indeed, once a samllchunks if processed the processed information must be written to disk
(instead of being saved in RAM).
For doing so, PyDmed provides a simple yet effective abstraction as illustrated by the following figure:
 The process of getting, e.g., heatmaps for WSIs has three components:
The process of getting, e.g., heatmaps for WSIs has three components:
- Dataloader: A dataloader (in this case
SlidingWindowDL) makes a stream of patches. - StreamCollector: This component recieves the stream of smallchunks generated by the dataloader. After processing one smallchunk, the StreamCollector hands over the “processed piece” (for instance the value of heatmap for that specific smallchunk) to the next step.
- StreamWriter: this component receives a stream of processed pieces and writes them on disk.
Please note that the three above components are done by different processes.
Specificly, the first and the third components make disk read/writes in a parallel way.
Therefore, the above scheme is very efficient.
PyDmed provides a straightforward way to make a customized StreamCollector.
For doing so, you can make a subclass from StreamCollector.
You are required to implement two methods:
- the function
process_pieceofstream: This function specifies how the outputs from the dataloader are processed. More precisely, given some return values from the dataloader, inprocess_pieceofstreamyou need to process the smallchunks and return a set ofProcessedPieces. - the fucntion
get_flag_finishcollecting: In this fucntion you should decide whether the whole processing has been finished. If it is finished, you have to return True and you have to return False otherwise. For the sliding window dataloader, you can leave the functionget_flag_finishcollectingas implemented in the below code.
The below box demonstrates how to build a StreamCollector that produces heatmaps for WSIs:
import pydmed.streamcollector
from pydmed.streamcollector import *
class HeatmapStreamCollector(StreamCollector):
def __init__(self, module_pipeline, device, *args, **kwargs):
#grab privates
self.module_pipeline = module_pipeline
self.device = device
#make other initial operations
self.module_pipeline.to(device)
self.module_pipeline.eval()
self.num_calls_to_getflagfinished = 0
super(HeatmapStreamCollector, self).__init__(*args, **kwargs)
@abstractmethod
def process_pieceofstream(self, retval_collatefunc):
x, list_patients, list_smallchunks = retval_collatefunc
with torch.no_grad():
netout = \
self.module_pipeline(x.to(self.device))#[32x1x7x7]
list_processedpiece = []
for n in range(netout.shape[0]):
tensor_piecen = netout[n,0,:,:].unsqueeze(0)
str_piecen = pydmed.extensions.wsi.Tensor3DtoPdmcsvrow(
tensor_piecen.detach().cpu().numpy(), list_smallchunks[n]
)
list_processedpiece.append(
ProcessedPiece(
data = str_piecen,\
source_smallchunk = list_smallchunks[n]
)
)
return list_processedpiece
@abstractmethod
def get_flag_finishcollecting(self):
self.num_calls_to_getflagfinished += 1
is_dl_running = self.lightdl.is_dl_running()
if((is_dl_running==False) and (self.lightdl.queue_lightdl.qsize()==0)):
return True
else:
return False
Pleae note that Tensor3DtoPdmcsvrow converts any numpy array of shape [CxHxW] to a string.
Indeed, str_piecen contains the values of [CxHxW] array as well as some other information needed to
make the C-dimensional feature map.
You can simply make an instance from HeatmapStreamCollector and call its start_collecting()
function:
streamcollector = HeatmapStreamCollector(
module_pipeline=model,
device = device,
lightdl = dl_forheatmap,
str_collectortype = "stream_to_file",
flag_visualizestats= False,
kwargs_streamwriter = {
"rootpath": "./Output/GeneratedHeatmaps/",
"fname_tosave":None,
"waiting_time_before_flush":3
}
)
streamcollector.start_collecting()
For each Patient, a separate csv file will be created in “./Output/GeneratedHeatmaps/”.
Getting heatmap for WISs takes several minutes.
After several minutes, all information to get heatmaps will be saved on those csv files.
Each line of those csv files includes the following information:
- (y,x): the position of the extracted smallchunk.
- (H,W): the size of the WSI at target level.
- level: the level from which the smallchunk is extracted.
- kernel size: the size of kernel.
- downsample in the level: the downsample (w.r.t level 0) for the target leve.
- C, H, W: the dimensions of the numpy array fed to
pydmed.extensions.wsi.Tensor3DtoPdmcsvrow. - content of the [CxHxW] array.
You can convert each csv file to a heatmap by calling one function as follows:
default_wsitoraster = pydmed.extensions.wsi.DefaultWSIxyWHvaltoRasterPoints()
fname_pdmcsv = "./Output/GeneratedHeatmaps/patient_1.csv"
np_heatmap = pydmed.extensions.wsi.pdmcsvtoarray(
fname_pdmcsv,
default_wsitoraster.func_WSIxyWHval_to_rasterpoints,
scale_upsampleraster = 1.0
)
You can increase the parameter scale_upsampleraster to increase the heatmap’s resolution.
Sample notebook to get heatmap is available at the following link: Link to sample notebook.
 |
 |